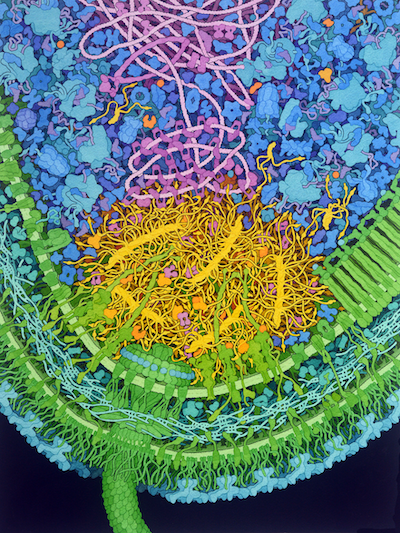
Molecular Landscapes by David S. Goodsell
Caulobacter Polar Microdomain, 2022
Acknowledgement: Illustration by David S. Goodsell and Keren Lasker, RCSB Protein Data Bank and Scripps Research. doi: 10.2210/rcsb_pdb/goodsell-gallery-046
The aquatic bacterium Caulobacter crescentus has a two-step life cycle: it is born as a free-swimming “swarmer” and differentiates into an immobile “stalked” cell. As part of this process, it populates the two ends of the cell (poles) with macromolecular complexes that regulate these two different life forms. These specialized polar complexes in turn guide construction of flagellar motors and pili in swarmers and a holdfast structure in stalked forms, and regulate replication and segregation of the DNA chromosome as the cell grows and divides. A distinctive microdomain, formed by the disordered protein PopZ, has the job of marking the poles and gathering these macromolecular complexes (“client” proteins) that have pole-specific functions. PopZ forms a condensate (yellow) that acts as a defined membraneless microdomain, excluding large cytoplasmic molecules like ribosomes from the pole, and selectively recruiting client proteins. In this illustration, the microdomain is interacting with several soluble clients involved in regulation (orange), several membrane-bound clients (yellow-green), and DNA binding proteins (magenta).
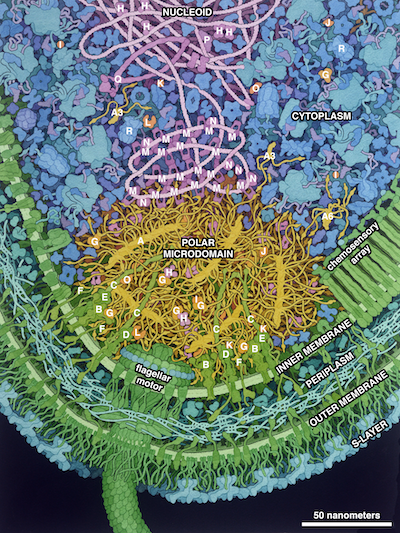
Key to the Illustration
PopZ: A, PopZ microfilament; A6, PopZ hexamer; A3, PopZ trimer. Clients: B, CckA; C, PodJ; D, PleC; E, DivL; F, ZitP; G, ChpT; H, CtrA; I, CpdR; J, RdcA; K, DivK; L, PleD; M, ParB; N, MipZ; O, PopA; P, ParA. Proteins related to the microdomain: Q, SMC (Structural Maintenance of Chromosomes); R, ClpXP.
For more information see:
Goodsell, DS Lasker, K (2023) Integrative visualization of the molecular structure of a cellular microdomain
Protein Science 32:e4577 doi: 10.1002/pro.4577
Curtis PD, Brun Y (2010) Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74(1):13-41.
Lasker K, Mann TH, Shapiro L (2016) An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Current Opinion in Microbiology 33:131-139.
Govers SK, Jacobs-Wagner C (2020) Caulobacter crescentus: model system extraordinaire. Curr Biol 30(19):R1151-1158.