|
Aconitase and Iron Regulatory Protein 1
Aconitase performs a reaction in the citric acid cycle, and moonlights as a regulatory protein
|
 |
Alcohol Dehydrogenase
Alcohol dehydrogenase detoxifies the ethanol we drink
|
 |
Alpha-amylase
Amylases digest starch to produce glucose
|
 |
ATP Synthase
ATP synthase links two rotary motors to generate ATP
|
 |
Bacteriorhodopsin
Bacteriorhodopsin pumps protons powered by green sunlight
|
 |
Beta-galactosidase
Beta-galactosidase is a powerful tool for genetic engineering of bacteria
|
 |
Biodegradable Plastic
Bacteria build biodegradable plastic that could be better for the environment
|
 |
Cellulases and Bioenergy
Powerful fungal enzymes break down cellulose during industrial production of ethanol from plant material.
|
 |
Citrate Synthase
Citrate synthase opens and closes around its substrates as part of the citric acid cycle
|
 |
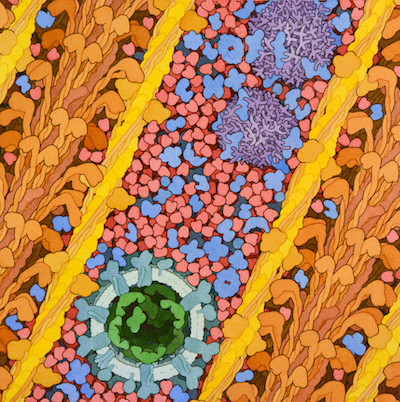
Citric Acid Cycle
Eight enzymes form a cyclic pathway for energy production and biosynthesis
|
 |
Complex I
A proton-pumping protein complex performs the first step of the respiratory electron transport chain
|
 |
Cytochrome bc1
A flow of electrons powers proton pumps in cellular respiration and photosynthesis
|
 |
Cytochrome c
Cytochrome c shuttles electrons during the production of cellular energy
|
 |
Cytochrome c Oxidase
Cytochrome oxidase extracts energy from food using oxygen
|
 |
Fatty Acid Synthase
Fatty acids are constructed in many sequential steps by a large protein complex
|
 |
GFP-like Proteins
GFP-like proteins found in nature or engineered in the laboratory now span every color of the rainbow
|
 |
Glycogen Phosphorylase
Glycogen phosphorylase releases sugar from its cellular storehouse
|
 |
Glycolytic Enzymes
The ten enzymes of glycolysis break down sugar in our diet
|
 |
Green Fluorescent Protein (GFP)
A tiny fluorescent protein from jellyfish has revolutionized cell biology
|
 |
Hypoxia-Inducible Factors
HIF-α is a molecular switch that responds to changing oxygen levels.
|
 |
Isocitrate Dehydrogenase
Atomic structures have revealed the catalytic steps of a citric acid cycle enzyme
|
 |
Lactate Dehydrogenase
Our cells temporarily build lactate when supplies of oxygen are low
|
 |
Luciferase
Organisms from fireflies to bacteria use luciferase to emit light
|
 |
Myoglobin
Myoglobin was the first protein to have its atomic structure determined, revealing how it stores oxygen in muscle cells.
|
 |
Photosynthetic Supercomplexes
Light is captured by huge supercomplexes of photosystems and antenna systems.
|
 |
Photosystem I
Photosystem I captures the energy in sunlight
|
 |
Phototropin
Phototrophins sense the level of blue light, allowing plants to respond to changing environmental conditions
|
 |
Pyruvate Dehydrogenase Complex
A huge molecular complex links three sequential reactions for energy production
|
 |
Pyruvate Kinase M2
Pyruvate kinases are the paradoxical gatekeepers for cancer cell metabolism and growth.
|
 |
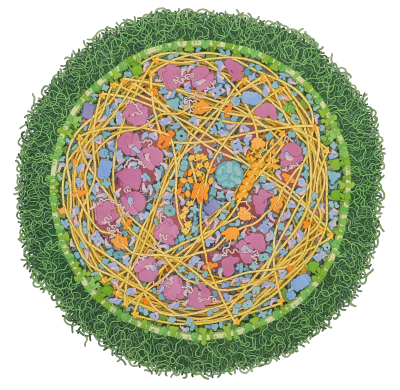
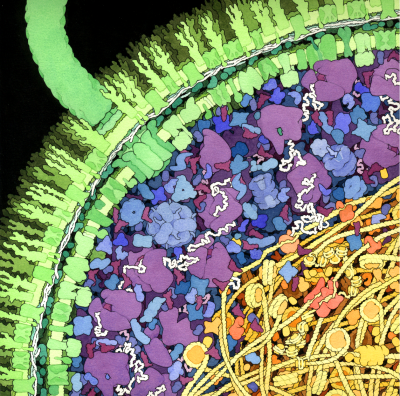
Respiratory Supercomplex
In our mitochondria, three electron-transport complexes assemble into a supercomplex.
|
 |
Rhodopsin
In our eyes, rhodopsin uses the molecule retinal to see light
|
 |
Trypsin
An activated serine amino acid in trypsin cleaves protein chains
|